Research Activities

- Computational Catalyst Design for Biomass Conversion
Non-food biomass is increasingly recognized as a valuable renewable energy resource, e.g., the non-food biomass in the USA alone was estimated to have the potential toproduce 3 billion barrels of liquidtransportation fuel annually. Cheap and efficient methodologies of converting biomass solids to liquid fuels on a large scale will not only help to solve for the energy needs of the word, but also will alleviate CO2 emission.
A promising pathway to generate liquid fuels from biomass in large scale is to design cheap, robust, and efficient catalysts (including homogeneous and heterogeneous catalysts and enzymes), based on the thorough understanding of the chemistry. Our group is interested in understanding the fundamental physical chemistry for biomass catalysts and also in designing cheap and efficient catalysts for converting biomass to liquid fuels.
- De Novo Drug Design for Cancer Therapeutics
We use computational chemistry methods to design molecular drugs for cancer therapeutics. We collaborate with experimentalists to test of validity of the designed drugs.
- Integrative Design of Polymer Nanospheres
Recently, we designed and synthesized (via the collaboration with Dr. Yin group at Sichuan University, China) a new class of polymeric nanospheres with the sizes of 30-120 nm in radius. The polymer nanospheres contain azo-benene functional dyes, and can change the shape under polarized light irradiation.
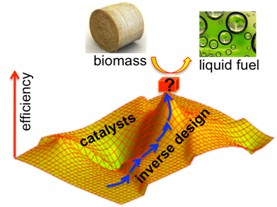
- Inverse Molecular Design in Tight-Binding Theory
Owing to the development in modern theoretical and computational chemistry (e.g., density functional theory), predicting molecular properties using accurate and efficient quantum chemistry methods becomes more and more practical. As a consequence, inverse molecular design approaches based on quantum chemistry have emerged as an attractive computational approach to take on the challenges in materials discovery.
Inverse molecular design is a general term describing strategies in molecular design that are in contrast to direct design methods. In direct design, a new molecule is proposed first, and then the molecular property is computed or measured to check its potential use. In contrast, inverse molecular design aims at searching for optimum points on hypersurfaces defining propertystructure relationships, and then mapping out the molecular structures atthe optimum points. Hence, using the idea of inverse molecular design could significantly enhance the efficiency and success rate of molecular design and save costs in materials discovery.
Our group has developed an inverse molecular design program (called LCAP-TB) on the basis of linear combination ofatomic potential (LCAP) in the tight-binding (TB) electronic structure theory. This approach allows us to search for optimum moleculesusing deterministic optimization algorithms. We have applied the LCAP-TB approaches to successfully design nonlinear optical materials and dye-sensitizers for solar cell applications. We are developing the TB-LCAP program for the design of biomass catalysts.


